Statistically Modelling the Spread of HIV
 Transmission Dynamics of HIV
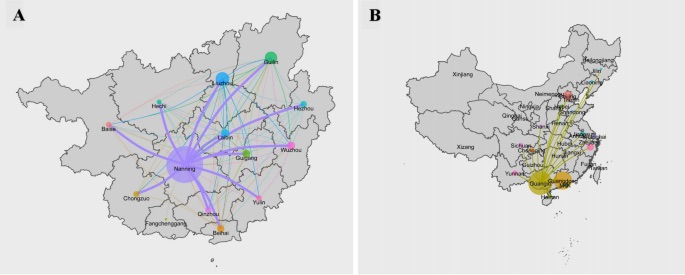
Transmission Dynamics of HIV
During the summer 2019 I joined Dr Oliver Ratmann for 2 months to conduct research on the spread of HIV. The project was funded by an EPSRC bursuary.
We performed a phylogeographic analysis to assess the geographic transmission rates. At the core of the project was a continuous-time Markov model. We worked with the software packages Beast to run the MCMC and FigTree to visualise the results. To tackle the issue of a high dimensional space with many negligible transmissions we applied Bayesian Stochastic Search Variable Selection (BSSVS). We quickly ran into an unexpected computational problem, as Beast did not allow the input of an existing tree, which shifted the project into the implementation of a function hardcoding the Beast input XML from an existing tree. The core reference for this project was this paper by Bbosa et al.